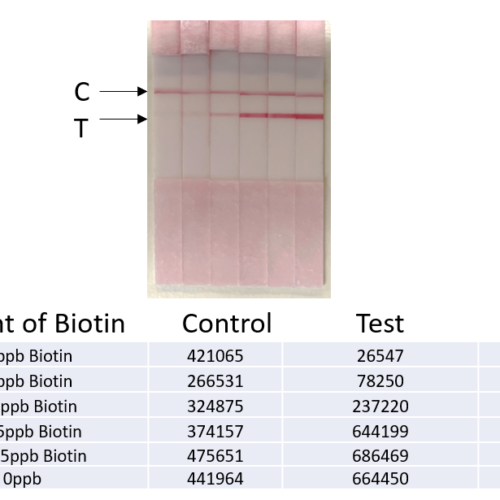
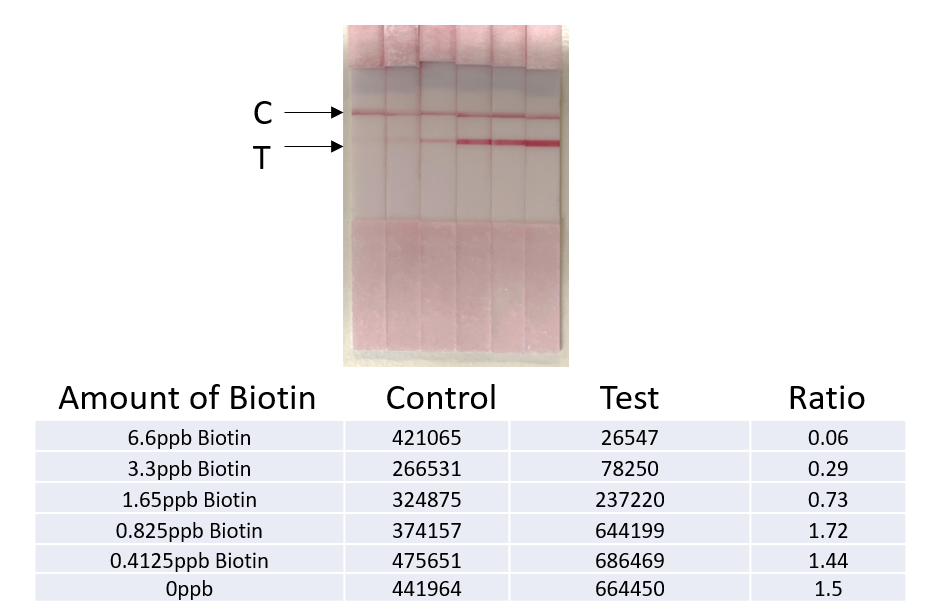
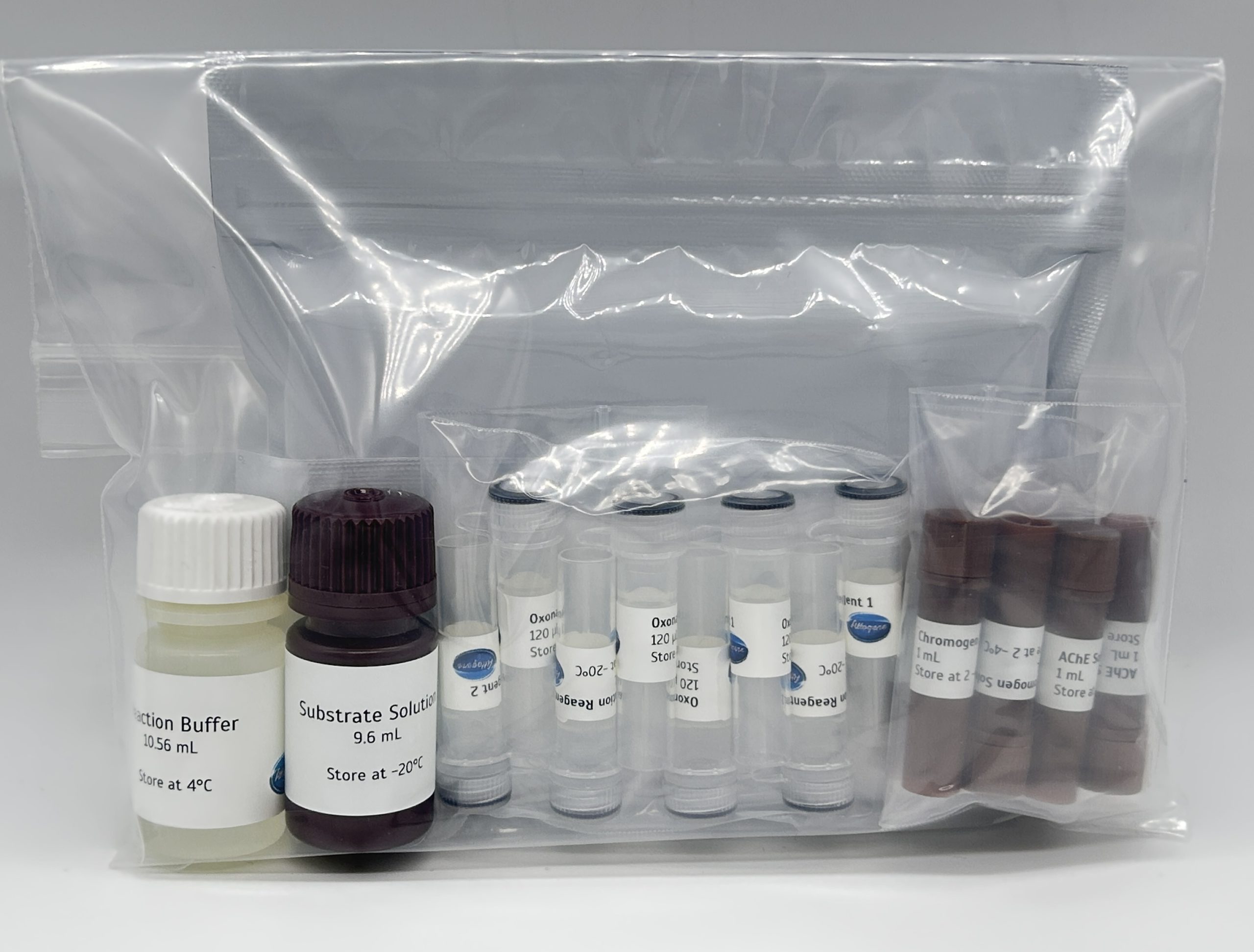
Legionella qPCR detection Kit
$376.25
In Stock & Ready to Ship
This kit is sufficient for 150 reactions:
- Mip region specific primer pair region
- 1.5mL PCR clean water
- SYBR Detection
Legionella pneumophila is a pathogenic bacterium primarily known for causing Legionnaires’ disease, a severe form of pneumonia. A crucial genetic marker for detecting this bacterium is the macrophage infectivity potentiator (Mip) gene. The Mip gene encodes a peptidyl-prolyl cis/trans isomerase enzyme, essential for bacterial entry, intracellular replication, and survival within host macrophages. Due to its high conservation and specificity within Legionella species, the Mip gene region serves as a reliable molecular target for PCR-based diagnostics and environmental monitoring.
Attogene’s qPCR kit for Legionella is designed for the in vitro analysis of the crucial genetic marker Mip. A gene-specific primer mix targeting the Mip region is provided for amplification and detection using SYBR Green dye on a qPCR instrument. samples are collected and processed to extract purified genomic DNA (gDNA). A reaction mixture is prepared using provided primers, SYBR Green master mix, and extracted gDNA samples as required. The assembled reaction is then loaded onto the qPCR instrument for amplification. The primer mix utilizes Taq polymerase to amplify the gene region of interest, with the SYBR Green dye binding specifically to double-stranded DNA during PCR amplification. This allows for real-time fluorescence detection across a wide range of qPCR platforms.
Recommended DNA isolation technique – inquire at sales@attogene.com for further details:
1. Sample Collection and Filtration:
- Collect water samples in sterile bottles, keeping them on ice or RT.
- Assemble the filtration system with a 0.2 µm or 0.45 µm membrane filter in the plastic membrane housing.
- Pass 100 mL to 1 L of water through the filter using a syringe.
- Carefully remove the membrane using sterile forceps and transfer it to a sterile 2.0 mL microcentrifuge tube.
- Store the membrane at -20°C (short-term) or -80°C (long-term) until DNA extraction.
2. Cell Lysis and DNA Extraction
- Cut the membrane into small pieces with a sterile scalpel and transfer to a 1.5 mL microcentrifuge tube.
- Add 500 µL of lysis buffer and 20 µL of Proteinase K.
- Incubate at 60°C for 30–60 minutes with occasional vertexing.
- Run through DNA extraction kit protocol or sonicate
3. Quantification and quality control
- Measure DNA concentration using a spectrophotometer.
- Use agarose gel electrophoresis to verify DNA integrity.
- Perform PCR amplification of internal control genes to confirm successful extraction.
You may also like…
Universal 23s PCR Amplification Kit (For cyanobacteria species identification)
$376.25- This kit is sufficient for 150 reactions:
- For characterizing cyanobacteria in environmental samples
- Use in combination with Attogene Algae DNA isolation kit
- Universal 23s PCR primers
- Perfect for Environmental DNA (eDNA) Characterization
Microcystin qPCR Detection Kit (real-time PCR kit for MycE Cluster)
$376.25This kit is sufficient for 150 reactions:
- Real time qPCR kit
- For screening microcystin gene cluster
- Use in combination with Attogene Algae DNA isolation kit
Compatible with automated systems.
Universal 16s PCR Amplification Kit (For cyanobacteria species identification)
$376.25- For Species identification of cyanobacteria in environmental samples
- Use in combination with Attogene Algae DNA isolation kit
- Perfect for Environmental DNA (eDNA) Characterization
Plant and Algae RNA Isolation Kit
$455.80This kit is sufficient for 100 RNA isolations based on:
- 200mg fresh plant
- 1ml algae culture
- 50mg dry seeds.
Compatible with automated systems.